The Microbial GAMUT Lab
Genomes And Metagenomes to Unravel Traits
Research Themes
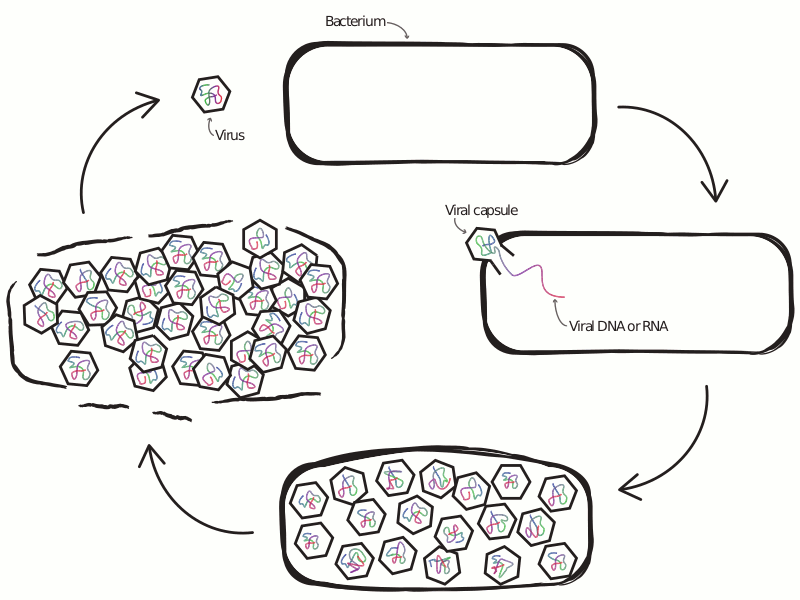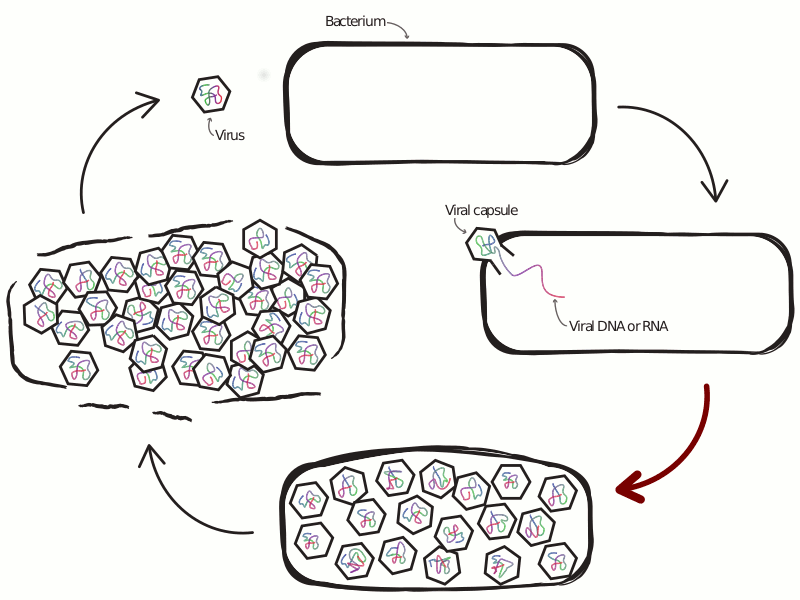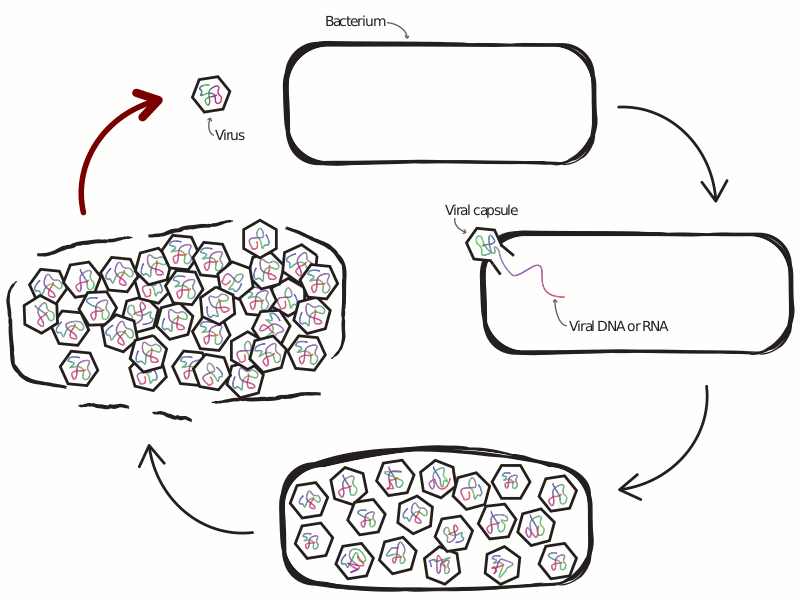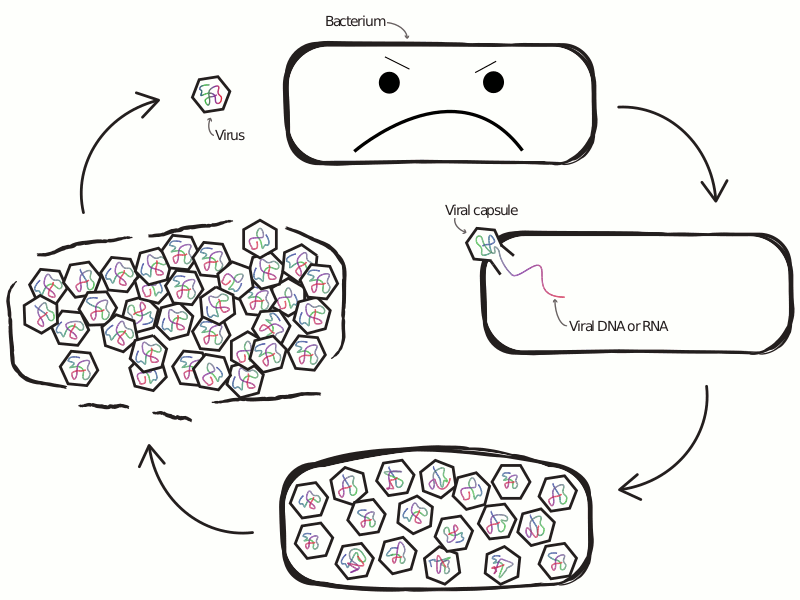
An Ongoing Battle Between Microbes and Their Viruses
Hey! Does this all seem a bit technical? Check out our article in Frontiers Young Minds for an explanation of CRISPR immunity that should be accessible to all ages
Viruses that infect microbes severely impact their hosts’ population and evolutionary dynamics. In an ecological context, these viruses lead to the release of important nutrients back into the environment and play a role in maintaining community-level diversity. In an evolutionary context, viruses drive the evolution of host immune strategy, often leading to iterative co-evolutionary dynamics. In the microbial world these two contexts are not distinct, with demographic and genetic changes occurring at similar rates, making any separation of scales infeasible. This is especially true at the interface of viral-host interactions, where the set of host defense and viral anti-defense strategies is diverse and fast-evolving. Extensive research has profiled the diversity of host and viral communities, but we know relatively little about the distribution of host defense and viral anti-defense strategies across environments.
What drives selection favoring a particular defense strategy? What new defense systems are still waiting to be discovered? How do defense systems coevolve with pathogens over time, and how does this coevolution feed back into the dynamics of host-virus interactions? The central theme of my previous and ongoing work is the application of tools from the fields of machine learning and complex systems science, alongside more traditional approaches from population genetics and theoretical ecology, to understand the ecology and evolution of antiviral defense strategies.
Trait-Based Analysis of Natural Microbial Communities
To-date efforts to functionally annotate metagenomes have largely been concerned with reconstructing the metabolic potential of communities. While microbial metabolism is incredibly diverse and an important aspect of functional ecology, microbes are more than just the metabolic pathways they encode. For example, even in the context of growth, extremely broad distributions of minimal doubling times are observed for microbial species with similar metabolic capacity (from under ten minutes to multiple days). I have built both genomic predictors and curated trait databases to help characterize the distribution of traits like maximal growth rate in natural communities.
Most recently I built a novel genomic estimator of maximal growth rate based on codon usage statistics that outperforms previous methods, including in a community context where I implemented a novel species abundance correction for metagenomes. In turn, I used this estimator to build a comprehensive database of over 200,000 growth rate estimates from genomes, metagenomes, and single-cell amplified genomes to survey growth potential across the range of prokaryotic diversity. The distribution of growth potentials suggested a natural divide between oligotrophs and coptiotrophs, and led me to propose a redefinition of these broad classes of microbes in terms of their selective environment. Additionally, my database revealed how culture collections, particularly of marine microbes, are strongly biased towards fast-growing organisms, with most environmentally-derived genomes on average having much slower predicted maximal growth rates than cultured isolates, illustrating that the current picture of microbial diversity is not only incomplete, but also highly skewed. I have implemented my growth-rate estimator in a user-friendly R package called gRodon.
Moving forward, I hope to leverage large phenotypic datasets to build improved genomic predictors for a variety of microbial traits - ultimately building an unbiased picture of microbial form and function in natural environments.
Background and Education
As of March 2020 I’m a Simons Foundation Postdoctoral Fellow in Marine Microbial Ecology in the Fuhrman Lab at USC.
Previously, I defended my PhD in Behavior, Ecology, Evolution, and Systematics (BEES) at the University of Maryland College Park coadvised by Philip LF Johnson (population genetics) and Bill Fagan (theoretical ecology). I was a COMBINE network science fellow (NSF), and before that a GAANN fellow in mathematical biology (US Dept. Ed.). My graduate training was broadly interdisciplinary with a heavy computational and theoretical emphasis, and I was lucky to have the opportunity to attend summer schools in Complex Systems Science at the Santa Fe Institute and in Microbial Diversity at the Woods Hole Marine Biological Laboratory.
I received a BA in mathematics and biology from Bard College in 2015 with a senior thesis advised by Bruce Robertson (biology) and Csilla Szabo (mathematics). As an undergraduate, I performed research under the supervision of faculty at Bard College, the Virginia Bioinformatics Institute, the Arizona State University Mathematics Department, and the Kavli Institute for Cosmological Physics at the University of Chicago.